-Search query
-Search result
Showing 1 - 50 of 351 items for (author: zimmer & j)
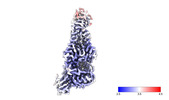
EMDB-40622: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer
Method: single particle / : Stephens Z, Zimmer J

EMDB-40623: 
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA
Method: single particle / : Stephens Z, Zimmer J

EMDB-40624: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP
Method: single particle / : Stephens Z, Zimmer J

PDB-8snc: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer
Method: single particle / : Stephens Z, Zimmer J

PDB-8snd: 
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA
Method: single particle / : Stephens Z, Zimmer J

PDB-8sne: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP
Method: single particle / : Stephens Z, Zimmer J

EMDB-40591: 
Xenopus laevis hyaluronan synthase 1
Method: single particle / : Gorniak I, Zimmer J

EMDB-40594: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
Method: single particle / : Gorniak I, Zimmer J

EMDB-40598: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state
Method: single particle / : Gorniak I, Zimmer J

PDB-8smn: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
Method: single particle / : Gorniak I, Zimmer J

PDB-8smp: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state
Method: single particle / : Gorniak I, Zimmer J
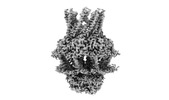
EMDB-41592: 
S. thermodepolymerans KpsMT(E151Q)-KpsE in complex with ATP
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41593: 
S. thermodepolymerans KpsMT-KpsE in complex with ADP:AlF4-
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41595: 
S. thermodepolymerans KpsM-KpsE in Apo 2 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41601: 
S. thermodepolymerans KpsMT-KpsE Apo 1
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41602: 
S. thermodepolymerans KpsM-KpsE in Glycolipid 2 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41626: 
S. thermodepolymerans KpsM-KpsE in Glycolipid 1 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41677: 
S. thermodepolymerans KpsMT-KpsE Apo 1 - consensus map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41678: 
S. thermodepolymerans KpsMT-KpsE Apo 1 - KpsT-focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41681: 
S. thermodepolymerans KpsMT(E151Q)-KpsE in complex with ATP - consensus map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41682: 
S. thermodepolymerans KpsMT(E151Q)-KpsE in complex with ATP - crown focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41697: 
S. thermodepolymerans KpsMT-KpsE in Apo 2 state - consensus map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41698: 
S. thermodepolymerans KpsMT-KpsE in Apo 2 state - KpsT focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41718: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 1 - consensus map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41720: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 1 - KpsT focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41721: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 1 - KpsM focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41771: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 2 - consensus map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41772: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 2 - KpsT focused map
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-41773: 
S. thermodepolymerans KpsMT-KpsE with bound glycolipid - state 2 - KpsM focused map
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tsh: 
S. thermodepolymerans KpsMT(E151Q)-KpsE in complex with ATP
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tsi: 
S. thermodepolymerans KpsMT-KpsE in complex with ADP:AlF4-
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tsl: 
S. thermodepolymerans KpsM-KpsE in Apo 2 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tsw: 
S. thermodepolymerans KpsMT-KpsE Apo 1
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tt3: 
S. thermodepolymerans KpsM-KpsE in Glycolipid 2 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

PDB-8tun: 
S. thermodepolymerans KpsM-KpsE in Glycolipid 1 state with rigid body fitted KpsT
Method: single particle / : Kuklewicz J, Zimmer J

EMDB-40477: 
KLHDC2 in complex with EloB and EloC
Method: single particle / : Digianantonio KM, Bekes M

PDB-8sh2: 
KLHDC2 in complex with EloB and EloC
Method: single particle / : Digianantonio KM, Bekes M

EMDB-15964: 
Subtomogram averaging of SARS-CoV-2 nsp3-4 delta Ubl1-Mac1 obtained from cryo-ET of cryo-FIB milled VeroE6 cells transfected with nsp3-4 delta Ubl1-Mac1
Method: subtomogram averaging / : Chlanda P, Zimmermann L

EMDB-15965: 
Subtomogram average of SARS-CoV-2 nsp3-4 delate Ubl1-Ubl2 obtained from cryo-ET of cryo-FIB milled VeroE6 cells transfected with nsp3-4 delta Ubl1-Ubl2
Method: subtomogram averaging / : Chlanda P, Zimmermann L

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
Method: single particle / : RAJAN KS, YONATH A

EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
